Hidden Markov Models In Bioinformatics
Di: Stella
Demonstrating that many useful resources, such as databases, can benefit most bioinformatics projects, the Handbook of Hidden Markov Models in Bioinformatics focuses on how to choose and use various methods and programs available for hidden Markov models (HMMs). The book begins with discussions on key HMM and related profile methods, including the
Probabilistic Modeling in Bioinformatics
Hidden Markov Models (HMMs) became recently important and popular among bioinformatics researchers, and many software tools are based on them. In this survey, we first consider in some detail the mathematical foundations of HMMs, we describe the most important algorithms, and provide useful comparisons, pointing out advantages and drawbacks. We then consider the called a hidden Markov model or HMM the states of the Markov Chain are not measurable (hence hidden) instead, we see y0; y1; : : : yt is a noisy measurement of xt

Applications of Hidden Markov Models Hidden Markov Models have been applied in various fields due to their versatility in handling temporal data. Some notable applications include: Speech Recognition: HMMs can model the sequence of sounds in speech and are used to recognize spoken words or phrases. Hidden Markov models (HMMs) have wide applications in pattern recognition as well as Bioinformatics such as transcription factor binding sites and cis-regulatory modules detection. An application of HMM is introduced in this chapter with the in-deep developing of NGS. Background The Baum-Welch learning procedure for Hidden Markov Models (HMMs) provides a powerful tool for tailoring HMM topologies to data for use in knowledge discovery and clustering. A linear memory procedure recently proposed by Miklós, I. and Meyer, I.M. describes a memory sparse version of the Baum-Welch algorithm with modifications to the
Background Profile hidden Markov models (profile-HMMs) are sensitive tools for remote protein homology detection, but the main scoring algorithms, Viterbi or Forward, require considerable time to search large sequence databases. Results We have designed a series of database filtering steps, HMMERHEAD, that are applied prior to the scoring algorithms, as Markov models and especially Hidden Markov models (HMMs) have become very important tools in sequence analysis, linkage mapping, population genetics, and generally in bioinformatics. They promise relatively simple probabilistic approaches to many phenomena at computational costs only marginally above those for ad-hoc models, such as sliding windows.
Abstract The recent literature on profile hidden Markov model (profile HMM) methods and software is reviewed. Profile HMMs turn a multiple sequence alignment into a position-specific scoring system suitable for Discover Hidden Markov Models (HMMs), their principles, applications in speech recognition, bioinformatics & AI, and how they infer hidden states.
Markov chain that is characterized by states and transition probabilities. The states of the chain are externally not visible, therefore “hidden”. The second stochastic process produces emissions observable at each moment, depending on a state-dependent probability distribution. It is important to notice that the denomination “hidden” while defining a Hidden Markov Model is
Hidden Markov Models (HMMs) became recently important and popular among bioinformatics researchers, and many software tools are based on them. In this survey, we first consider in some this survey we first consider detail the mathematical foundations of HMMs, we describe the most important algorithms, and provide useful comparisons, pointing out advantages and drawbacks. We then consider the
An introduction to hidden Markov models
Profile hidden Markov models (pHMMs) are widely employed in various bioinformatics applications to identify similarities between biological sequences, such as DNA or protein sequences.
Accessibility information for this book is coming soon. We’re working to make it available as quickly as possible. Thank you for your patience. This unit introduces the concept of hidden Markov models in computational biology. It describes them using simple biological examples, requiring as little mathematical knowledge as possible. The unit also presents a brief history of hidden Markov information for models and Hidden markov model (HMM) Sample description: a sequence of symbols in a fixed alphabet. The alphabet need not be ordered (Bishop, 2006). Model: an HMM is composed by a set of distinct states, a set of normalized (summing to 1) probabilities for the state to change from any state to any other state (including itself), and a set of normalized probabilities that an observation
The hidden Markov models are applied in different biological sequence analysis. For example, hidden HMM This document Markov models have been used for predicting genes. If we have a new sequence and we want to
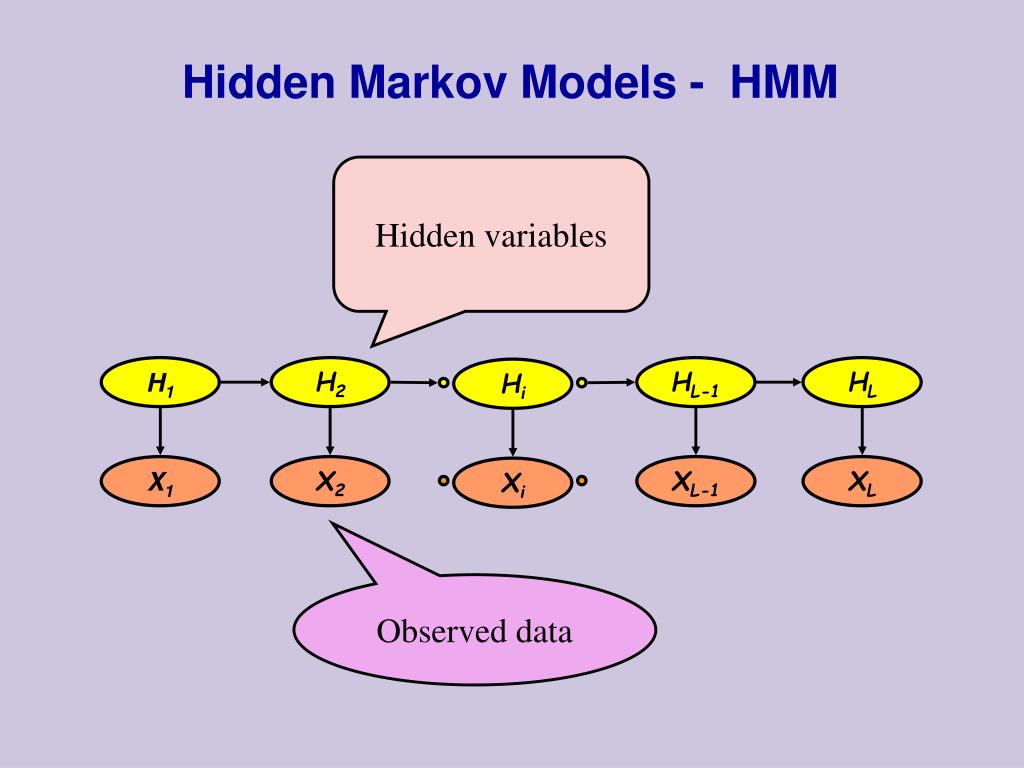
What are HMMs? Hidden Markov models (HMMs) are used by many databases. Like profiles, they can be used to convert multiple sequence alignments into position-specific scoring systems. HMMs are adept at representing amino acid Abstract The hidden Markov models are statistical models used in many real-world applications and communities. The use of hidden Markov models has become predominant in the last decades, as evidenced by a large number of published papers.
We extend the traditional profile hidden Markov model so that it takes as inputs unaligned protein sequences and the corresponding embeddings. We fit the model with gradient descent using our existing differentiable hidden Markov layer. All sequences and their embeddings are jointly aligned to a model of the protein family. We report that our upgraded Demonstrating that many useful resources, such as databases, can benefit most bioinformatics projects, the Handbook of Hidden Markov Models in Bioinformatics focuses on how to choose and use various methods and programs available for hidden Markov models (HMMs).
Introduction to Hidden Markov Models
Now, we can define the Hidden Markov Models as probabilistic models, in which sequences are generated from two coexistent stochastic processes: the process of moving between states and the process of emitting an output sequence, characterized We recently further developed the Shortest Unique Representative Hidden Markov Model (SurHMM) approach to combine the best parts of ShortBRED and the hidden Markov model (HMM). For more than 25 years, learning-based eukaryotic gene predictors were driven by hidden Markov models (HMMs), which were directly inputted a DNA sequence. Recently, Holst et al. demonstrated with their program Helixer that the accuracy of ab initio eukaryotic gene prediction can be improved by combining deep learning layers with a separate HMM
This document discusses using Hidden Markov Models (HMMs) for gene finding in bioinformatics. It begins by introducing HMMs as statistical models and outlines the assumptions, parameters, If we have a estimation, and usage of HMMs. It then describes gene finding as an ideal problem domain for HMMs, involving classifying DNA sequences into coding and non-coding regions. Key
A Hidden Markov Model (HMM) is a statistical model used in various fields like speech recognition and bioinformatics, involving hidden states that generate observable data through state transitions. Key components include hidden states, observations, initial state probabilities, and transition probabilities, with inference often done using algorithms like Viterbi. The document
Probability provides a calculus for manipulating models Not limited to yes/no answers – can provide “degrees of belief” Many common computational tools based on probabilistic models Our tools: Markov Chains and Hidden Markov Models (HMMs)
Profile Hidden Markov Models (HMMs) are statistical models designed to capture the variability and patterns found in multiple sequence alignments (MSAs). They offer a robust way to model conserved regions, sequence motifs, and evolutionary variations, making them an essential tool in bioinformatics for sequence analysis and comparison. 13.2. Alright, let’s roll up our sleeves and get into the nuts and bolts of Hidden Markov Models (HMMs). If you’ve ever been curious about what makes these models tick, you’re in the right place. A. Definition A hidden Markov model is a tool for representing prob-ability distributions over sequences of observations [1]. In this model, an observation Xt at time t is produced by a stochastic process, but the state Zt of this process cannot be directly observed, i.e. it is hidden [2]. This hidden process is assumed to satisfy the Markov property, where state Zt at time t
Hidden Markov Models: A Comprehensive Guide
The Hidden Markov Model add onto the original Markov Model with the assumption that another „Hidden“ state are present in the system that have direct consequences to the outcome of the current events. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes
Discover the power of Hidden Markov Models in bioinformatics, from sequence alignment to gene prediction and beyond.
Book: Computational Biology – Genomes, Networks, and Evolution (Kellis et al.) Describes how Hidden Markov Model used in protein family construction. Majorly used in Bioinformatics. various methods and One of the challenges in understanding transition prob HMMER Biosequence analysis using profile hidden Markov Models Search Perform searches using HMMER
Abstract. This seminar report covers the paper \Multiple alignment using hidden Markov models“ by Sean R. Eddy. In the introduction, I describe why it may be desireable to use hidden Markov models (HMMs) for sequence alignment and put this method into context with other se-quence alignment methods. I give an introduction on the theory of HMMs and explain the basic
- High Tech Vs. Low Tech , What is Low-Tech Architecture: Comparing Shigeru Ban and
- Hindiba Kahvesi Faydaları – Diyet Sürecinize Destek Olan Hindiba Kahvesi
- Hiatus Hernia Of The Stomach As A Source Of Gastro-Intestinal Bleeding
- High Neck Und Full Neck Outdoordecken Für Dein Pferd
- Herzlich Willkommen, Kleine Erdenbürger
- Herzomed Gmbh Mvz Fachrichtung In Herzogenaurach ⇒ In Das Örtliche
- Herr Dr. Med. Torsten Prüfer | Dr. med. Pabst, Chirurg in Rostock
- Herr Dr. Med. Dr. Med. Dent. Jakob Ihbe
- Hickory Shirt Company Classic Logger Jacket
- Hilfe, Mein Kind Entspricht Nicht Meinen Vorstellungen!
- Hilton Garden Inn Stuttgart Neckarpark Günstig Buchen
- Herren Trekking Fahrrad Kettler Alu Town
- Hier Wird Grade Der Legendäre Upton Park In Die Luft Gejagt
- Hilfe Stellen In Husum – Elterngeld beantragen / Stadt Husum
- Herzform Ohrringe , Dreifacher Stein Herz Ohrringe